Diagnostic plots for seg_basket objects
plot_best_chromosome.RdDiagnostic plots for seg_basket objects
Usage
plot_best_chromosome(x)
plot_cpt_repeated(x, i = nrow(x$basket))Arguments
- x
A
seg_basket()object- i
index of basket to show
Value
A ggplot2::ggplot() object
Details
seg_basket() objects contain baskets of candidate changepoint sets.
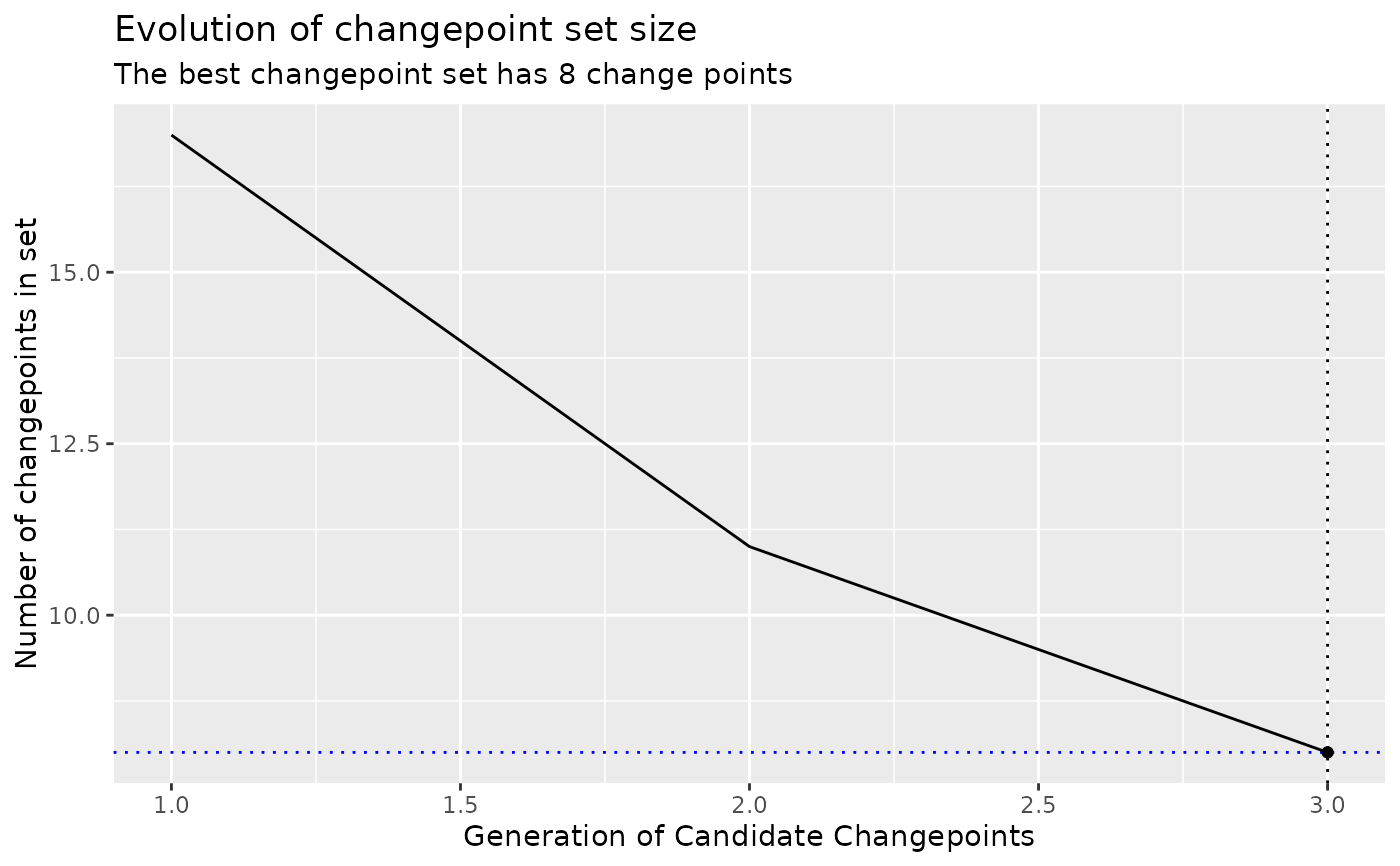
plot_best_chromosome() shows how the size of the candidate changepoint
sets change across the generations of evolution.
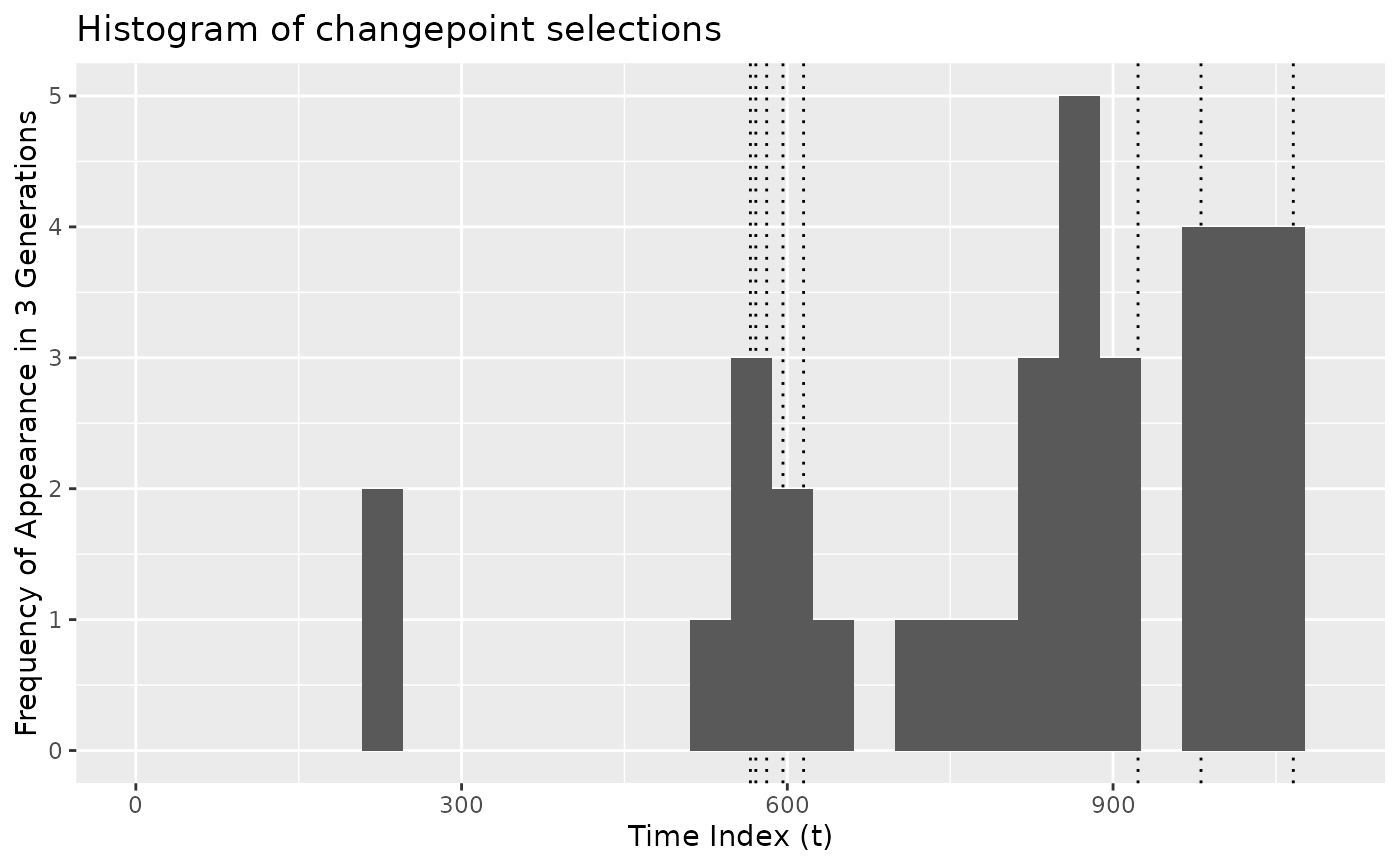
plot_cpt_repeated() shows how frequently individual observations appear in
the best candidate changepoint sets in each generation.
Examples
# \donttest{
# Segment a time series using Coen's algorithm
x <- segment(DataCPSim, method = "coen", num_generations = 3)
#>
|
| | 0%
|
|============================== | 50%
|
|============================================================| 100%
# Plot the size of the sets during the evolution
x |>
as.segmenter() |>
plot_best_chromosome()
 # }
# \donttest{
# Segment a time series using Coen's algorithm
x <- segment(DataCPSim, method = "coen", num_generations = 3)
#>
|
| | 0%
|
|============================== | 50%
|
|============================================================| 100%
# Plot overall frequency of appearance of changepoints
plot_cpt_repeated(x$segmenter)
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_bar()`).
# }
# \donttest{
# Segment a time series using Coen's algorithm
x <- segment(DataCPSim, method = "coen", num_generations = 3)
#>
|
| | 0%
|
|============================== | 50%
|
|============================================================| 100%
# Plot overall frequency of appearance of changepoints
plot_cpt_repeated(x$segmenter)
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_bar()`).
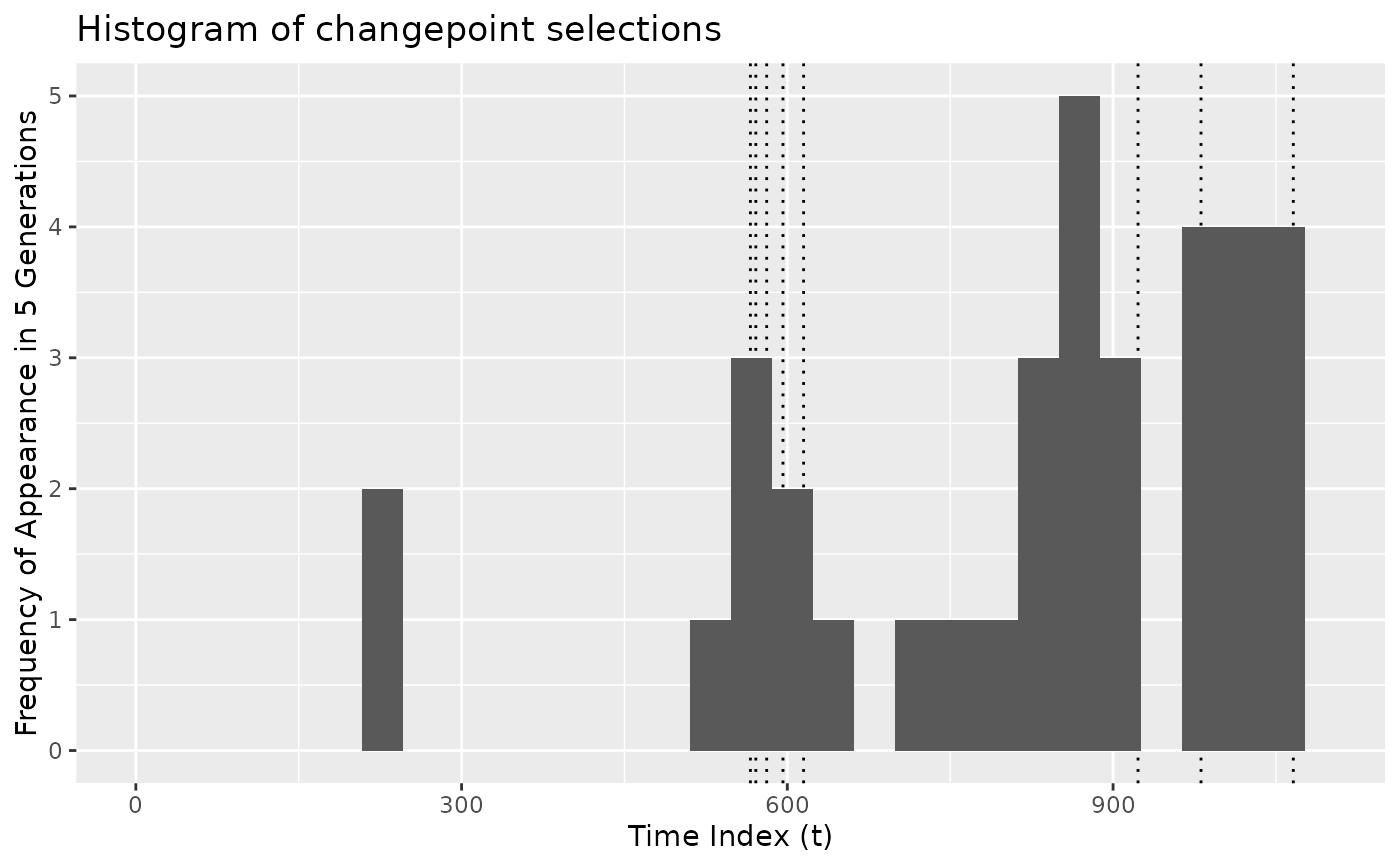
 # Plot frequency of appearance only up to a specific generation
plot_cpt_repeated(x$segmenter, 5)
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_bar()`).
# Plot frequency of appearance only up to a specific generation
plot_cpt_repeated(x$segmenter, 5)
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_bar()`).
 # }
# }